One could easily argue that further posts demonstrating the evidence for common descent from genomics are superfluous given that the case has already been made beyond reasonable doubt. However, the point of these posts is to show that given multiple independent lines of evidence converge on the same conclusion, the special creationist argument is clearly shown to be iterated special pleading.
The power of the evidence from endogenous retroviral elements is that they are clearly alien to the human genome as they are undeniably evidence of ancient retroviral infection that became integrated into the germ line, and subsequently inherited by descendant species. The odds of exactly the same retrovirus integrating into the germ line at exactly the same place in the genomes of related species is billions to one against just for a single retroviral element. Given that there are multiple such examples in human and ape DNA, the chance becomes so remote as to be negligible.
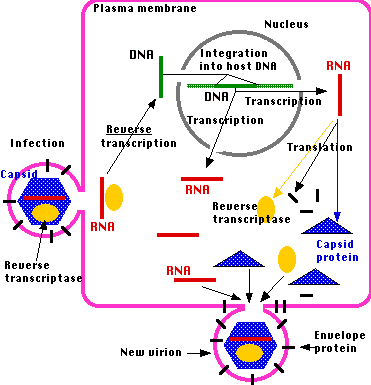
Retroviruses are RNA viruses that reproduce intracellularly by using their reverse transcriptase enzyme to produce a DNA copy of their genome which is then inserted into the host genome. Once the DNA copy is part of the host cell, the host cell genetic replication machinery produces new copies of the retrovirus.
The power of the evidence from endogenous retroviral elements is that they are clearly alien to the human genome as they are undeniably evidence of ancient retroviral infection that became integrated into the germ line, and subsequently inherited by descendant species. The odds of exactly the same retrovirus integrating into the germ line at exactly the same place in the genomes of related species is billions to one against just for a single retroviral element. Given that there are multiple such examples in human and ape DNA, the chance becomes so remote as to be negligible.
Retroviruses are RNA viruses that reproduce intracellularly by using their reverse transcriptase enzyme to produce a DNA copy of their genome which is then inserted into the host genome. Once the DNA copy is part of the host cell, the host cell genetic replication machinery produces new copies of the retrovirus.
Retroviral life cycle. Source
If the retrovirus integrates into the host's germ line, then it can be passed down to the next generation. When that happens, it becomes an endogenous retrovirus. As the DNA copy does not produce material essential to the well-being of the cell (as one would expect given its viral origins) it will eventually become inactivated by mutation. The presence of endogenous viral elements in an organism's genome is proof of a prior retroviral infection. When two related organisms share the same ERV at exactly the same place in the genome, we have powerful evidence that these organisms share a common ancestor in which the original viral infection took place and was then passsed down to the descendant species.
While pseudogenes and retrotransposons are 'indigenous' to the species in question for want of a better term, ERV data is unarguably alien, presence of an ancient retroviral infection that became integrated into the germ line of the animal, and passed down the generations. Therefore, the presence of identical retroviral elements at the same position in human and ape genomes strongly suggests infection and integration of the retroviral element in a species ancestral to human and ape.
Common descent would predict that if a species was infected by a retrovirus which became fixed in the genome, all the descendant species should also inherit this ERV. The probability that multiple species have independently been infected by the same retrovirus at the same point in their genomes is quite remote. The respected virologist John Coffin notes:
“Because the site of integration in the genome, which comprises some three billion base pairs in humans, is essentially random, the presence of an ancient provirus at exactly the same position in different, but related, species cannot occur by chance, but must be a consequence of integration into the DNA of a common ancestor of all the species that contain it. It evolution of retroviruses follows, therefore, that we can infer what viruses were present millions of years ago by examining the distribution of endogenous proviruses in modern species.” [1]
ERVs are in addition very tightly integrated into the genome, making it extremely hard for them to be neatly excised without leaving behind remnants:
Therefore, an ERV locus shared by two or more species is descended from a single integration event and is proof that the species share a common ancestor into whose germ line the original integration took place. Furthermore, integrated proviruses are extremely stable: there is no mechanism for removing proviruses precisely from the genome, without leaving behind a solo LTR or deleting chromosomal DNA. The distribution of an ERV among related species also reflects the age of the provirus: older loci are found among widely divergent species, whereas younger proviruses are limited to more closely related species. [2]
It’s hard to get any more definite than that. The chances of the same retrovirus inserting itself into the same location in the genomes of a human and a chimpanzee is around 1 in 3,000,000,000. If we have multiple ERVs found in the same places in related species, then the odds of this occurring by chance become so remote as to be impossible.
When ERV element distribution in humans and primates is examined, what we see is entirely what common descent predicts. Johnson, in a paper coauthored with Welkin Johnson showed that ERV elements can be used to construct primate evolutionary family trees. This hinges on the principle that ERVs, once integrated into the germ line will be inherited, and therefore of use as markers of inheritance. They used human endogenous retroviruses (HERVs) and found that these HERVs are:
When ERV element distribution in humans and primates is examined, what we see is entirely what common descent predicts. Johnson, in a paper coauthored with Welkin Johnson showed that ERV elements can be used to construct primate evolutionary family trees. This hinges on the principle that ERVs, once integrated into the germ line will be inherited, and therefore of use as markers of inheritance. They used human endogenous retroviruses (HERVs) and found that these HERVs are:
the result of integration events that took place between 5 and 50 million years ago, as indicated by the distribution of specific proviruses at the same integration sites (or loci) among related species. The evolution of primates has been the subject of intense study for well over a century, providing a well established phylogenetic consensus with which to compare and evaluate the performance of ERVs as phylogenetic markers. [3]
The idea behind this is fairly simple. An ERV element should not be under positive selection as it is of no use to the organism, and will eventually accumulate random mutations. The longer the time between the divergence of the two lines leading to the modern speies, the more mutations will accumulate in these ERV elements. From this data, an evolutionary family tree can be constructed.
ERV elements differ from pseudogenes and retrotransposons in that they have three sources of information that allow evolutionary family trees to be constructed:
- The distribution of ERVs among related species
- Accumulated mutations in ERVs, allowing an estimate of genetic distance
- Sequence divergence between the LTRs at each end of the ERV, which is a source of information unique to endogenous retroviruses.
The odds of this distribution of ERV elements occuring by chance is remote. The vertebrate genome is huge, and retroviral integration is random, making the odds of identical ERV integration at the same place in multiple genomes unlikely:
Therefore, an ERV locus shared by two or more species is descended from a single integration event and is proof that the species share a common ancestor into whose germ line the original integration took place. Furthermore, integrated proviruses are extremely stable: there is no mechanism for removing proviruses precisely from the genome, without leaving behind a solo LTR or deleting chromosomal DNA. The distribution of an ERV among related species also reflects the age of the provirus: older loci are found among widely divergent species, whereas younger proviruses are limited to more closely related species. [4]
The second point has been addressed previously, and need not be covered again. The final point is one unique to ERVs. At each end of the ERV is a sequence known as a LTR, or Long Terminal Repeat. The mechanics of reverse transcription mean that both LTRs will be identical when the ERV integrates into the genome. Johnson and Coffin note:
Furthermore, both clusters are predicted to have similar branching patterns as determined by the phylogenetic history of the host species, with similar branch lengths. Thus, each tree displays two estimates of host phylogeny, both of which are derived from the evolution of an initially identical sequence. As we shall see, deviation of actual trees from this prediction provides a powerful means of testing the assumptions and detecting events other than neutral accumulation of mutations in the evolutionary history of a species. [5]
Johnson and Coffin looked at the distribution of ERVs in the primate genetic material analused, and found:
Three of the loci, HERV-KC4, HERV-KHML6.17, and RTVL-Ia, were detectable in the genomes of OWMs and hominoids, but not New World monkeys, and therefore integrated into the germ line of a common ancestor of the Old World lineages. HERV-K18, RTVL-Ha, and RTVL-Hb were found exclusively in humans, gorillas, chimpanzees, and bonobos, and thus are consistent with a gorilla/chimpanzee/human clade. None of the loci was detected in New World monkeys. [6]
This is perfectly explained by common descent. To reiterate an “ERV locus shared by two or more species is descended from a single integration event and is proof that the species share a common ancestor into whose germ line the original integration took place.” Johnson and Coffin found many loci shared by these primate species, some shared only by humans, chimps, bonobos and gorillas, some shared only by old world monkeys and hominoids (humans and great apes). This data is consistent with an evolutionary origin of these species, but impossible to explain by special creation.
Most of the ERVs analysed produced phylogenetic trees consistent with expectation. Their conclusions:
The HERVs analyzed above include six unlinked loci, representing five unrelated HERV sequence families. Except where noted, these sequences gave trees that were consistent with the well established phylogeny of the old world primates, including OWMs, apes, and humans… Phylogenetic analysis using HERV LTR sequences gives rise to trees with a predictable topology, on which is superimposed the phylogeny of the host taxa, and allows ready detection of conversion events. [7]
Other studies show that humans and primates share ERVs in a way consistent with common descent. Barbulescu et al showed that many human ERVs of the HERV-K class (present in humans, apes and old world monkeys) are unique to humans:
Two proviruses, HERV-K105 and HERV-K110/HERV-K18 were detected in both humans and apes. HERV-K110 was present in humans, chimpanzees, bonobos and gorillas but not in the orangutan. Thus, this provirus formed after orangutans diverged from the lineage leading to gorillas, chimpanzees, bonobos and humans, but before the latter species separated from each other. HERV-K105 was detected in humans, chimpanzees and bonobos, but not in gorillas or the orang-utan. The preintegration site, however, could not be detected in gorillas or orang-utans using several different primers based on the human sequences that flank this provirus. It is therefore unclear from this analysis whether this provirus formed after gorillas diverged from the human–chimpanzee–bonobo lineage, or if it formed earlier but was subsequently deleted in one or more lineages leading to modern apes. It is clear that at least one full-length HERV-K provirus in the human genome today has persisted since before humans, chimpanzees, bonobos and gorillas separated during evolution, while at least eight formed after humans diverged from the extant apes. [8]
Belshaw et al, looking at the long-term reinfection of the human genome by ERVs note that:
Within humans, the most recently active ERVs are members of the HERV-K (HML2) family. This family first integrated into the genome of the common ancestor of humans and Old World monkeys at least 30 million years ago, and it contains >12 elements that have integrated since the divergence of humans and chimpanzees, as well as at least two that are polymorphic among humans…This recent activity makes this family ideal for distinguishing between the alternative mechanisms of proliferation. [9]
The pattern of HERV-K elements, shown below, demonstrates just how powerful the ERV evidence is in demonstrating human-ape common ancestry, as well as confirming the standard evolutionary history of primates. Again, ERVs are remnants of ancient viral infection. They are not native to humans or primates, but bear witness to ancient retroviral infection. The odds of multiple identical HERV elements integrating into primate and human DNA in exactly the right places to simulate common descent are so low as to be effectively zero.
This is exactly what we see when we examine human and primate genomes - multiple ERVs inserting at the same place in their respective genomes. More importantly, the pattern of insertion of these ERVs matches the standard evolutionary family tree. Medstrand and Mager [10] examined the pattern of insertions of a particular class of endogenous retroviruses, the HERV-K family. Thirty-seven ERV fragments were aligned into clusters based on sequence divergence. When they compared this with primate genomic data, they found that the clusters with greater divergence were also found in Old World monkeys and apes, while those with a lesser amount of divergence were found only among gorillas and chimpanzees. The cluster with the least amount of divergence was found only in the human genome.
The pattern of HERV-K element insertion shown above once again matches what common descent would predict:
In general, LTR sequences of clusters 1 to 5 were first identified in Old World monkey and gibbon DNAs, whereas LTRs of cluster 8 first appeared in DNAs of gorilla and chimpanzee. For example, the AF001550 LTR of cluster 3 is not present in Old World monkeys but is present in gibbon and all higher primates. In contrast, the AC003023 cluster 8 LTR is found only in chimpanzee and human, indicating a more recent integration, Initial results with primers flanking three of the integrated LTRs of cluster 9 resulted in the expected amplification products in human DNA but not in any of the other primate DNAs, To demonstrate that sequences of cluster 9 were unique to human DNA, primers flanking the other six identified LTRs of this cluster, including the full-length HERV-K10 element, were used in the amplification of primate DNA. Indeed, all were detected only in human DNA, indicating that sequences derived from this cluster integrated after the divergence of the human lineage from the great apes. [11]
It is this nesting of HERV clusters, in a way according with what common descent predicts that makes this powerful evidence for common descent. ERVs are evidence of ancient retroviral infection, and the presence of the same ERV at the same place in related species is as Coffin states prima facie evidence for an ancient retroviral infection in the ancestor of both species. When we multiply the number of ERVs that have integrated in the same place in many primate genomes, but do so in the pattern above, the case for common descent based solely on ERV inclusions becomes overwhelming.
Special creationist attacks don't even address the main evidence ERVs provide for common descent
Attempts in our community to explain away the evidence for common descent from shared ERV elements betray a considerable lack of familiarity with the subject. For example, David Burges, science editor of The Testimony claims that maybe the retroviral elements will turn out to have a function after all - and reaches the point where he ends up making unprovable assertions that the Fall somehow warped the genomes of humans and animals:
For instance, the human genome contains sequences of what appear to be inactive retrovirus genomes incorporated in the same specific places in the genomes of everyone. The hypothesis is that these are the results of random insertions into the genome of a common human ancestor, since the same apparent 'ancient' retrovirus sequences are located in exactly the same equivalent place, between the same genes, in the equivalent chromosomes of some primates, including bonobos, chimps, and gorillas (although not in those of others such as orang-utans). [12]
LTR—gag—pol—env—LTR
LTR: long terminal repeat. They are DNA sequences that are involved in the insertion of the retroviral genome into host DNA.
gag: group specific antigen. This codes for retroviral structural proteins
pol: polymerase. This codes for reverse transcriptase, protease and integrase
env: envelope. This codes for the retroviral coat proteins.
It is hard not to miss the decayed remnants of retroviral infection when we see the decayed remains of retroviruses littering the genome. Furthermore, we know that these ERVS are alien to the vertebrate genome because of codon bias. As ERV researcher Abigail Smith notes:
Viruses, including retroviruses, including endogenous retroviruses, dont speak the same language as humans. Sure they use A, T/U, C, G nucleotides in codons, coding for amino acids that make proteins.
But viruses and humans dont speak this language with the same accent. Its called codon bias, or codon-pair bias.
Several codons can code for the same amino acid. For instance, GCU, GCC, GCA, GCG all code for Alanine. So, in the human genome, you would expect ~25% of all the Alanines to use GCU, ~25% to use GCC, ~25% to use GCA, and ~25% to use GCG, right? Nope. For some reason, the human genome prefers GCC over GCG. Four times as many Alanines are coded by GCC as GCG. Humans have a GCC ‘accent’.
Viruses have their own codon ‘accents’ as well. And even though the differences should, theoretically, mean nothing (an Alanine is an Alanine is an Alanine), one way we can attenuate viruses to make better vaccines is to force them to use codons they dont like.
Thus one of the ways we know ERVs are a later addition to the human genome, and didnt originate in the human genome, is that retroviruses have a different codon ‘accent’ than humans. They don't fit. [13]
Furthermore, we have evidence of retroviral insertion in real-time. Since ERVs have been found in every vertebrate whose genome has been examined, it is not unreasonable to look for both evidence of active ERV infection and examples of ERV inclusions in other related species. This is in fact what we see. Take the subject of active retroviral infection. The koala genome is currently being colonised by the koala retrovirus (KoRV). Research [14] has shown that:
KoRV is present, at variable copy number, in the germline of all koalas found in Queensland, but that animals from some areas of southern Australia lack the provirus. Most notably, KoRV appears completely absent from koalas on Kangaroo Island off the coast of South Australia. This island was stocked with koalas in the early part of the twentieth century and has remained essentially isolated since then; it appears most likely that the small founding population was entirely free of KoRV. Tarlington et al. suggest that an ongoing process of infection and endogenization is now occurring, spreading from a focus in northern Australia that quite possibly initiated within the last 100 to 200 years. [15]
This by the way is not something that is of particular immediate benefit to the koala:
KoRV appears to be associated with the fatal lymphomas that kill many captive animals . It may also be immunosuppressive, thereby contributing to the chlamydial infections that afflict many koalas. [16]
There is no doubt that what we see in our genome is the result of ancient retroviral infection, and as I pointed out earlier, respected virologist John Coffin notes:
As a rule, they entered the germ line before the origin of the species, and are therefore found in every individual at the same genomic location. Because of their long residence in the host DNA, they have accumulated mutations and other genetic damage that cause them to be inactive, and, in general, they cannot be activated to yield infectious virus. Because the site of integration in the genome, which comprises some three billion base pairs in humans, is essentially random, the presence of an ancient provirus at exactly the same position in different, but related, species cannot occur by chance, but must be a consequence of integration into the DNA of a common ancestor of all the species that contain it. [17]
Therefore, when Burges continues by asserting:
Time may tell whether these are actually ancient retroviral sequences or whether they have some other function.
he is incorrect. These are remains of ancient retroviral sequences. Human and primate genomes are littered with the remains of ancient retroviral infection, and as he motes, the presence of an ancient proviral element at the same place in different but related species is evidence of common ancestry.
Furthermore, Burges, like almost all special creationist critics of evolution, has confused ERV elements - such as LTRs and the retroviral genes gag, pol, and env - with a fully functional ERV. Occasionally, evolution co-opts ERV elements but this is not the same thing as a fully functional ERV which would simply produce more retroviruses. One does not want a fully functional ERV in the genome producing retroviruses, given the disease-causing potential of retroviruses such as HIV. Burges' lack of understanding of retrovirology and genomics is obvious when he asserts:
The argument that common gene sequences must mean common ancestry (as opposed to common design) is an assertion which disregards alternative explanations.
Remember, that the argument starts fro, the premise that all primates descended from common ancestors.
Burges ignores the fact that the common design alternative has been examined and discarded. There is here a subtle shift from shared identical proviral elements to common gene sequences, a rhetorical shift which frankly is evasive. Even then, this does not help Burges. As I've mentioned before, the considerable functional redundancy in the genetic code means that for even a small 100 amino acid protein, there are something like 10^49 possible ways to code for exactly the same protein. Common descent would predict that the gene sequences for all life today would be clustered together in such a way that the phylogenetic tree constructed from the genomic data would be consonant with the standard phylogenetic tree. Conversely, under a model of special creation, there would be no reason for humans and chimps to have gene sequences closely related. However, this is exactly what we see. The special creation hypothesis is therefore rejected.
Secondary co-option of ERV elements does not invalidate their use to demonstrate common descent
There is plenty of evidence to show that the genome can co-opt retroviral elements for another function. For example, there is strong evidence that the development of the mammalian placenta was contingent on syncytin, a retroviral envelope protein that had inserted itself into the mammalian line millions of years ago. [18] There is no doubt that this is a viral gene, not a vertebrate gene. Furthermore, syncytin is in the same place in the genomes of humans, apes and monkeys. [19] Science writer Carl Zimmer comments:
Viruses have insinuated themselves into the genome of our ancestors for hundreds of millions of years. They typically have gotten there by infecting eggs or sperm, inserting their own DNA into ours. There are 100,000 known fragments of viruses in the human genome, making up over 8% of our DNA. Most of this virus DNA has been hit by so many mutations that it’s nothing but baggage our species carries along from one generation to the next. Yet there are some viral genes that still make proteins in our bodies. Syncytin appeared to be a hugely important one to our own biology. Originally, syncytin allowed viruses to fuse host cells together so they could spread from one cell to another. Now the protein allowed babies to fuse to their mothers. [20]
The fact that it is functional is not the issue - it is performing a completely different function to what it used to to when it was a viral gene. Rather, we have an identical gene completely alien to the mammalian genome in the same place in primate genomes, which is proof that primates - including humans - have a common ancestor which had been originally infected by the retrovirus which carried the syncytin gene.
Special creationist attempts to rebut this unarguable evidence for common descent invariably miss the point, and fixate on the co-option of elements of the endogenous retrovirus by the host genome, and ignore the fact that it is the presence of identical retroviral elements (many of which have not been co-opted by the host genome) at the same place in the genomes of related species which provides the evidence for common descent.
Casey Luskin, a lawyer and former geology student who works for the Discovery Institute, an intelligent design advocacy group has argued:
Casey Luskin, a lawyer and former geology student who works for the Discovery Institute, an intelligent design advocacy group has argued:
In his "29+ Evidences for Macroevolution" on TalkOrigins, Douglas Theobald claims that "Endogenous retroviruses provide yet another example of molecular sequence evidence for universal common descent." The presumption behind his argument is that endogenous retroviruses (ERVs) are functionless stretches of "junk" DNA that persist because they are "selfish"--but they have no function for the organism. If we find the same ERVs in the same genetic loci in different species of primates, Theobald concludes they document common ancestry. But what if ERVs do perform important genetic functions? Even theistic evolutionist Francis Collins acknowledges that genetic similarity "alone does not, of course, prove a common ancestor" because a designer could have "used successful design principles over and over again." (The Language of God, pg. 134.) The force of Theobald's argument thus depends upon the premise that ERVs are selfish genetic "junk" that do not necessarily perform any useful function for their host.In contrast, ID proponents would predict function for ERVs. This isn't because ID has an inherent quarrel with common descent--it doesn't. Rather, ID predicts function because the basis for ID's predictions is observations of how intelligent agents design things, and intelligent agents tend to design objects that perform some kind of function. As William Dembski wrote in 1998, "If, on the other hand, organisms are designed, we expect DNA, as much as possible, to exhibit function." It seems that the expectations of ID are turning out to be right.
A recent 2008 paper, "Retroviral promoters in the human genome," in the journal Bioinformatics (Vol. 24(14):1563--1567 (2008)) discusses the fact that "Endogenous retrovirus (ERV) elements have been shown to contribute promoter sequences that can initiate transcription of adjacent human genes. However, the extent to which retroviral sequences initiate transcription within the human genome is currently unknown." The article thus "analyzed genome sequence and high-throughput expression data to systematically evaluate the presence of retroviral promoters in the human genome."
The results were striking:
We report the existence of 51,197 ERV-derived promoter sequences that initiate transcription within the human genome, including 1743 cases where transcription is initiated from ERV sequences that are located in gene proximal promoter or 5' untranslated regions (UTRs).
[...]
Our analysis revealed that retroviral sequences in the human genome encode tens-of-thousands of active promoters; transcribed ERV sequences correspond to 1.16% of the human genome sequence and PET tags that capture transcripts initiated from ERVs cover 22.4% of the genome. These data suggest that ERVs may regulate human transcription on a large scale.
(Andrew B. Conley, Jittima Piriyapongsa and I. King Jordan, "Retroviral promoters in the human genome," Bioinformatics, Vol. 24(14):1563--1567 (2008).)
Darwinists who labeled ERVs as a form of "selfish" and "junk" DNA have been chasing explanations down a blind alley. It should be stated that the authors do not deviate from the neo-Darwinian paradigm, putting the obligatory evolutionary spin on the data. They claim that it's a possibility that some of the transcribed ERVs are "not functionally significantl," exposing that even in the face of this compelling contrary data, it is difficult for many Darwinists to let go of their seductive but science-stopping "junk-DNA" paradigm.
Luskin uses an inept bait-and-switch here:
Theobald concludes they document common ancestry. But what if ERVs do perform important genetic functions? Even theistic evolutionist Francis Collins acknowledges that genetic similarity "alone does not, of course, prove a common ancestor" because a designer could have "used successful design principles over and over again." (The Language of God, pg. 134.)
Similarity alone does not prove common descent, but ERVs are not 'indigenous' to the species. They are clearly viral in origin, and evidence of prior retroviral infection. Luskin fails to mention this point because as mentioned earlier, the presence of identical viral material at the same loci is consistent with infection in a common ancestor.
The question of function is irrelevant - the human genome has co-opted retrotransposons and ERV promoters to perform function over time. However, this was never their intended function in the first place - retrotransposons are mobile genomic elements whose presence is evidence of an earlier copying and insertion event and primarily exist solely to copy themselves, while ERVs are as mentioned evidence of ancient retroviral infection.
The question of function is irrelevant - the human genome has co-opted retrotransposons and ERV promoters to perform function over time. However, this was never their intended function in the first place - retrotransposons are mobile genomic elements whose presence is evidence of an earlier copying and insertion event and primarily exist solely to copy themselves, while ERVs are as mentioned evidence of ancient retroviral infection.
The force of Theobald's argument thus depends upon the premise that ERVs are selfish genetic "junk" that do not necessarily perform any useful function for their host. In contrast, ID proponents would predict function for ERVs. This isn't because ID has an inherent quarrel with common descent--it doesn't.
The force of Theobald's argument has been shamelessly misrepresented by Luskin. This is what Theobald said:
Endogenous retroviruses provide yet another example of molecular sequence evidence for universal common descent. Endogenous retroviruses are molecular remnants of a past parasitic viral infection. Occasionally, copies of a retrovirus genome are found in its host's genome, and these retroviral gene copies are called endogenous retroviral sequences. Retroviruses (like the AIDS virus or HTLV1, which causes a form of leukemia) make a DNA copy of their own viral genome and insert it into their host's genome. If this happens to a germ line cell (i.e. the sperm or egg cells) the retroviral DNA will be inherited by descendants of the host. Again, this process is rare and fairly random, so finding retrogenes in identical chromosomal positions of two different species indicates common ancestry. (Emphasis mine)
Theobald's argument is not primarily based on 'junk' DNA, but the simple fact that shared identical ERV genomic material in two species is evidence of common descent.
Conclusion
I became involved in cancer research, and in the early 1980s, read avidly to inform myself of dramatic developments in the genetics of cancers. It was then that I came across oncogenic retroviruses. These are a subtype of virus that had a cunning mode of propagating themselves, and they were revolutionizing our understanding of how cancers developed. They brought to light a class of genes known as oncogenes. I struggled to assimilate the deluge of data, totally focused on cancer biology, my professional interest. But to my enormous surprise, I was following a continuous track which led to the point where I found myself reading in the area of evolutionary biology.
Retroviruses provided a way of demonstrating that many cancers are produced from a single abnormal cell. Counter-intuitive though it may seem, the billions of cells that may populate a tumour are the descendants of a single ancestral cell, so cancers are said to be monoclonal. And, almost unbelievably, retroviruses provided a way of showing that multiple species may be derived from a single progenitor species (indeed, ultimately from a single cell). Such related taxa of organisms are said to be monophyletic.
As I read, I found that a large variety of genetic markers established both the monoclonal nature of tumours on the one hand, and the monophyletic nature of groups of species on the other. Humans, chimps, gorillas and orang-utans, for example, share millions of genetic markers that show – unambiguously – that the four species share a common history. The genetic principles applicable to cancer (or immunology or microbiology or whatever) and evolutionary phylogenetics were the same, thoroughly established and non-controversial.
Once again, nothing in medicine makes sense except in the light of evolution, particularly the pattern of retroviral element distribution we see in humans and the great apes.
References
10. Medstrand P, Mager DL “Human-Specific Integrations of the HERV-K Endogenous Retrovirus Family” J Virol (1998) 72(12):9781-9787
11. ibid, p 9784
14. Tarlington RE, Meers J, Young PR: Retroviral invasion of the koala genome. Nature 2006, 442:79-81.
15. Stoye JP “Koala retrovirus: a genome invasion in real time” Genome Biology 2006, 7:241-3
17. Coffin JM “Evolution of Retroviruses: Fossils in our DNA” Proceedings of the American Philosophical Society (2004) 148:264-280
19. Mallet F et al "The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology" Proc Natl Acad Sci USA (2004) 191:1731-1736
20. Zimmer C "Mammals Made by Viruses" The Loom Feb 14th 2012
21. Finlay G "Human Evolution: Genes, Genealogies, and Phylogenies" BioLogos Blog May 27 2014
1. Coffin JM “Evolution of Retroviruses: Fossils in our DNA” Proceedings of the American Philosophical Society (2004) 148:264-280
2. Johnson WE Coffin JM Constructing primate phylogenies from ancient retrovirus sequences Proc. Natl. Acad. Sci USA (1999) 96:10254-10260
3. ibid p 10254
4. ibid p 10255
5. Johnson WE Coffin JM op cit p 10255-10256
6. ibid p 10256
7. ibid p 10259
8. Barbulescu M et al “Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans” Current Biology (1999) 9:861-868
9. Belshaw R et al “Long-term reinfection of the human genome by endogenous retroviruses” Proc. Natl. Acad. Sci. USA. (2004) 101:4894-4899
11. ibid, p 9784
12. Burges D "Is Theistic Evolution Compatible With Faith in God's Word?" The Testimony (2014) 84:143-147
13. Smith A "Claim 8– ERVs were Specially Created portions of the human genome before The Fall" Oct 6 2009
15. Stoye JP “Koala retrovirus: a genome invasion in real time” Genome Biology 2006, 7:241-3
16. ibid, p 243
18. McCoy J.M. et al "Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis" Nature (200) 403:785-789
19. Mallet F et al "The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology" Proc Natl Acad Sci USA (2004) 191:1731-1736
20. Zimmer C "Mammals Made by Viruses" The Loom Feb 14th 2012
21. Finlay G "Human Evolution: Genes, Genealogies, and Phylogenies" BioLogos Blog May 27 2014