The Genomic Evidence for Common Descent: 4. Retrotransposons
The evidence from nuclear gene phylogenies and shared identical pseudogenes alone is enough to confirm human-ape common ancestry beyond reasonable doubt, but the evidence from comparative genomics does not end there. Retrotransposons, mobile genetic elements that copy and paste themselves randomly throughout the genome provide another line of evidence for common descent. Unlike pseudogenes, retrotransposons are essentially selfish genetic material, existing solely to propagate itself throughout its genomic host. The presence of identical retrotransposon material at the same place in the genomes of related species is prima facie evidence that those species shared a common ancestor in which the retrotransposition event first took place. Once again, there is simply no credible special creationist explanation for their existence.
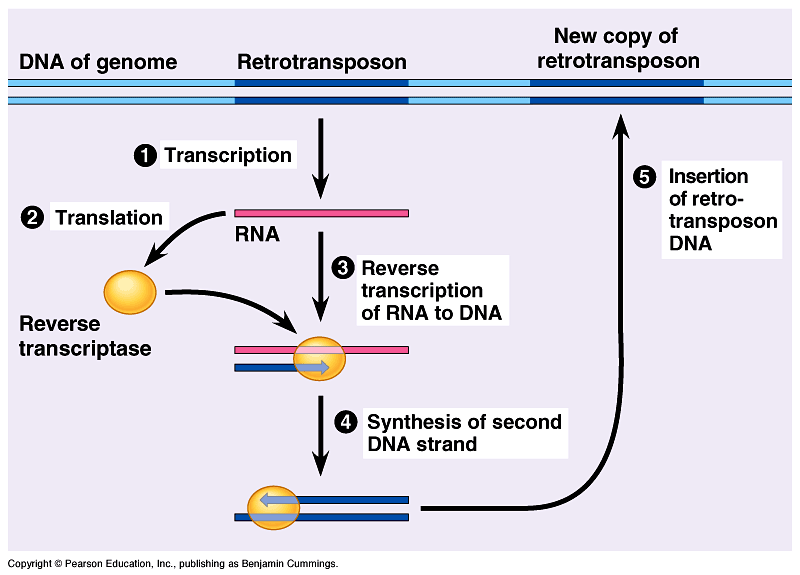
Retrotransposons are a class of genetic element that copy and paste themselves throughout the genome; unlike DNA transposons, they copy themselves into an RNA intermediate which is inserted randomly into the genome via the process of reverse transcription.
They are divided into two main classes: LTR retrotransposons, which share many similarities with endogenous retroviruses, and non-LTR retrotransposons which are subdivided into two further groups:
- SINEs - Short Interspersed Elements
- LINEs - Long Interspersed Elements
Unlike LINEs, SINEs do not code for reverse transcriptase, the enzyme used to convert an RNA sequence to DNA to permit insertion into the genome, and therefore need other transposable elements to assist in retrotransposition. As retrotransposons copy and paste themselves through the genome, over time they will accumulate and form a large part of the host genome. Around 45% of the human genome consists of these mobile genetic parasites. [1]
 |
| Source |
Uncommonly, retrotransposons can be co-opted by the host genome to perform a useful function, but generally, their benefit at best is neutral, and sometimes can be deleterious. As Robert Trivers and Austin Burt note:
Due to their abundance, transposable elements are almost guaranteed to have profound effects on their hosts. About half of our own genome is derived from transposable elements. And, in addition to transposition itself, these elements can cause a bewildering array of chromosomal rearrangements. As with other types of mutations, some fraction of these insertions and rearrangements will be beneficial to the host and positively selected. Transposable elements may thus be important sources of mutation that would not occur by other means. And transposable elements can also be domesticated (rarely) by the host-as we shall see, just such an event was critical to the evolution of the vertebrate immune system. These beneficial effects notwithstanding, transposable elements are still best considered as parasites, not as host adaptations or mutualists. (Emphasis mine) [2]
Parasite is an excellent word to use to describe these transposable elements. Ignoring the tiny fraction that has been co-opted, almost all of the retrotransposon complement is largely dead metabolic weight. They are also directly linked to genetic disease [3-6], a point that respected evolutionary biologist John Avise notes:
Mobile elements have the potential to cause human diseases by several mechanisms. When a mobile element inserts into a host genome, it normally does so at random with respect to whether or not its impact at the landing site will harm the host. If it happens to land in an exon, it can disrupt the reading frame of a functional gene with disastrous consequences. If it jumps into an intron or an intron-exon boundary, it may cause problems by altering how a gene product is spliced during RNA processing. If it inserts into a gene’s regulatory region, it can also cause serious mischief. The potential for harm by such insertional mutagenesis is great. It has been estimated, for example, that an L1 or Alu mobile element newly inserts somewhere in the genome in about 1–2% and 5%, respectively, of human births. Another problem is that when a mobile element lands in a functional gene, genetic instabilities are sometimes observed that result in deleted portions of the recipient locus. Several genetic disorders have been traced to genomic deletions associated with de novo insertions of mobile elements. Finally, mobile elements (or their immobile descendents that previously accumulated in the human genome) can also cause genomic disruptions via non-allelic homologous recombination. Serious metabolic disorders can result. [7]
While this does not prove common descent (though it completely overturns any argument that retrotransposons were specially created parts of the human genome), the presence of shared retrotransposons at identical sections of genomes in related species confirms common descent. A classic example is a study that confirmed hippos and whales share a common ancestor, using retrotransposon data:
Analysis of the KM14, HIP4, HIP24, and AF loci show that hippopotamuses and cetaceans form a monophyletic group that excludes ruminants. In each case, the CHR-1 SINE was inserted into the genome of a common ancestor of these species, as confirmed by hybridization experiments with probes specific for CHR-1 and its flanking region...On closer examination, it was revealed that the CHR-1 SINE was integrated into a common ancestor of hippopotamuses and whales…(indicating a sister-group relationship between these taxa), whereas a MER unit was integrated into the genome of a common ancestor of ruminants only. [8]
The presence of identical retrotransposons at the same place in the genomes of related species is prima facie evidence for common descent. As mentioned, these are mobile genetic elements that are copied from elsewhere and insert themselves randomly into other places in the genome. The odds of the same SINE or LINE inserting at the same place in the genomes of related species is vanishingly small, making common descent the only credible explanation for the pattern of retrotransposon distribution seen. As Nikaido et al point out:
SINES and LINEs are virtually unique and irreversible mutations...which is well documented with primate Alu (SINE) sequences. During the last 10 years, one of us (N.O.) has studied several hundred SINE loci, but he has never observed any occurrence of independent SINE insertions among species at identical genomic positions (i.e., between the same two nucleotides). Because the probability that a SINE / LINE will be lost once it has been inserted into the genome is extremely small, and the probability that the same SINE / LINE will be inserted independently into an identical region in the genomes of two different taxa is also very small, the probability that homoplasy will obscure phylogenetic relationships is, for all practical purposes, zero...Therefore, one can reconstruct phylogenetic trees with high confidence by considering multiple independent SINE / LINE insertion events that define given nodes in a tree. (Emphasis mine) [9]
As the presence of retrotransposons in the genome is evidence of prior copying and pasting of that genetic element, if we see the same retrotransposon in exactly the same place in the genome of two related species, then we have two options:
- Purely by chance, the same retrotransposon inserted itself into exactly the same place in human and ape genomes
- The retrotransposon was first inserted in the genome of the common ancestor of human and ape, and then inherited by the descendant species.
The odds of independent identical retrotransposon insertion at identical places in human and ape genomes is remote. If we have an example where seven retrotransposons are found in exactly the same places in human and chimpanzee genomes, then the chances are so remote as to be impossible. This is exactly what we have found. In the alpha haemoglobin cluster of humans and chimpanzees. Researchers have found that "[i]n each case, with the exception of minor sequence differences, the identical Alu repeat is located at identical sites in the human and chimpanzee genomes." [10] In fact, by looking for the presence of retrotransposons in primate genomic data, we have been able to construct a reliable evolutionary family tree that is consistent with the accepted tree based on morphological data.
 |
| Source: Gene (2007) 390:39-51 |
The use of the unique pattern of retrotransposon data has real-world applications in allowing researchers to identify - for example - what sort of primates a carnivore eats based on the genetic material detected in its faeces. It also allows accurate identification of products seized in the illegal wildlife trade. [11] It is also worth noting that this approach can be used to identify humans. [12] Special creationists who accept the result of a paternity test should, to be consistent, accept this technique when it confirms common descent of humans and apes.
References
1. Lander ES, Linton LM, Birren B, et al. "Initial sequencing and analysis of the human genome" Nature (2001) 409: 860–921
2. Trivers R, Burt A "Genes in Conflict: The Biology of Selfish Genetic Elements" (2009: Harvard University Press) p 229
3. Ostertag E.M. et al "SVA Elements Are Nonautonomous Retrotransposons that Cause Disease in Humans" Am J Hum Genet (2003) 73:1444-14514. Callinan, P. and Batzer, M.A. (2006) Retrotransposable elements and human disease. In Genome and Disease. Genome Dynamics (Vol. 1) (Volff, J., ed.), pp. 104–115, Karger
5. Crow, Mary K. "Long interspersed nuclear elements (LINE-1): potential triggers of systemic autoimmune disease." Autoimmunity (2009) 43: 7-16.
6. Schneider, Anna M., et al. "Roles of retrotransposons in benign and malignant hematologic disease." Cellscience (209) 6:121
7. Avise J.C., "Footprints of nonsentient design inside the human genome" Proc. Natl. Acad. Sci. USA (2010) 107:8969-8976
8. Mikaido M et al "Phylogenetic relationships among cetartiodactyls based on insertions of short and long interpersed elements: Hippopotamuses are the closest extant relatives of whales" Proc. Natl. Acad. Sci. USA (1999) 96:10261-10266
9. ibid, p 10264
10. Sawada I. et al "Evolution of alu family repeats since the divergence of human and chimpanzee" Journal of Molecular Evolution (1985) 22:316-322
11. Scott W. Herke S.W. et al "A SINE-based dichotomous key for primate identification" Gene (2007) 390:39-51
12. Novick G.E. et al. "Polymorphic human specific Alu insertions as markers for human identification." Electrophoresis (1995) 16:1596-1601.
10. Sawada I. et al "Evolution of alu family repeats since the divergence of human and chimpanzee" Journal of Molecular Evolution (1985) 22:316-322
11. Scott W. Herke S.W. et al "A SINE-based dichotomous key for primate identification" Gene (2007) 390:39-51
12. Novick G.E. et al. "Polymorphic human specific Alu insertions as markers for human identification." Electrophoresis (1995) 16:1596-1601.


Comments
Post a Comment